GlioSeq® - NGS Panel for Brain Tumors

GlioSeq Advantage
- Simultaneous sequencing and detection of genomic alterations
- Small Sample Size
- Fast Turnaround Time
Test Details
- GlioSeq v3 is utilizing an amplification-based targeted next generation sequencing of DNA and mRNA
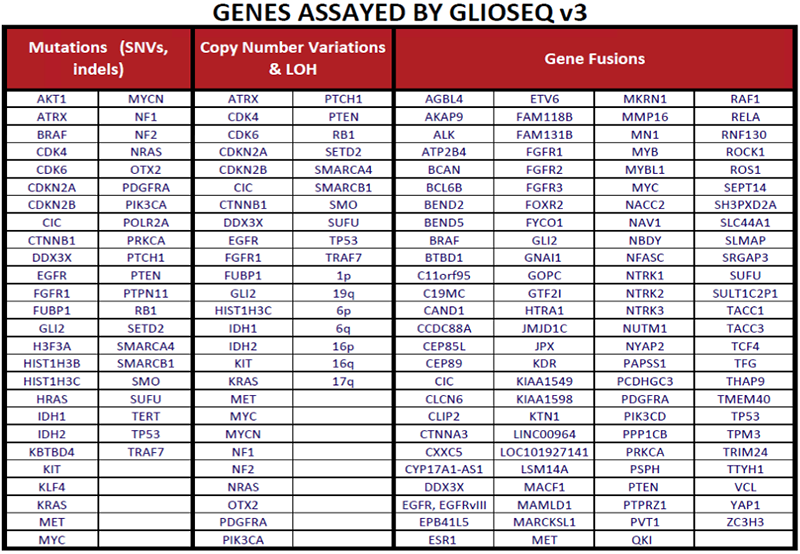
- GlioSeq version 3 next generation sequencing analysis is performed to detect base substitutions (SNVs) and small insertions/deletions in targeted regions of 47 key brain tumors genes, for copy number changes in 43 genes, and structural alterations involving 103 genes including BRAF, NTRK1-3, FGFR3, YAP1, RELA, EGFR (EGFRvIII), and others
- It can be performed on small stereotactic brain biopsies and resected FFPE tumor specimens
- It has fast turnaround time (7-10 days)
- GlioSeq aids in the diagnosis of both adult and pediatric CNS tumors, including glioma, medullosblastoma, meningioma, and ependymoma
Genes Assayed by GlioSeq
Background
Molecular markers are used to support and enhance the diagnosis, prognosis and treatment of adult and pediatric CNS tumors. The GlioSeq test identifies genetic alterations that are relevant to different CNS tumors subtypes and grades. [1, 2]
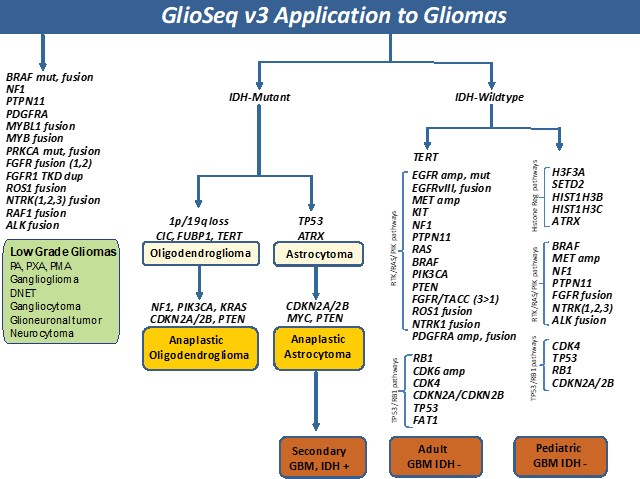
Low grade gliomas (WHO Grade I-II), including pilocytic astrocytoma, pilomyxoid astrocytoma, ganglioglioma, pleomorphic xanthoastrocytoma, and other neuronal and neuroglial tumors often harbor mutations or gene fusions in BRAF, PDGFRA, PTPN11, FGFR family genes, NTRK2, RAF1, and ALK.[3-5] Neurofibromatosis type 1 associated pilocytic astrocytoma characteristically harbor mutations and/or loss in NF1 gene, resulting in bi-allelic inactivation of the gene and frequently seen in the younger age group with multiple tumors located retro-orbitally, along the optic tract, and base of the brain.[6] In addition, genetic alterations in NTRK3, NTRK1, MYBL1, MYB, FGFR1, and RAF1 genes are commonly seen in pediatric low-grade gliomas.[2, 3, 7, 8]
Diffusely infiltrative Gliomas (WHO grade II-III) were classified into three glioma subtypes based on histopathologic features, molecular alterations, and clinical behavior.[9] Oligodendrogliomas harbor mutations in IDH1 and IDH2 genes, 1p/19q co-deletion, TERT promoter mutation and alterations in CIC, and FUBP1 genes. Infiltrating astrocytoma harbor IDH1/2, TP53, and ATRX mutations. Both can progress to a higher-grade glioma by acquiring additional genomic alterations in the RTK-RAS-PI3K pathway genes. A subset of lower grade gliomas do not harbor IDH mutations but have genetic alterations similar to high grade gliomas (WHO grade IV) and considered to be a precursor to IDH-mutant (secondary) GBMs.[9-11]
Primary GBMs (WHO grade IV) are IDH wild-type and harbor a number of genetic alterations that lead to dysregulation of critical signaling pathways including i) receptor tyrosine kinase (RTK)/RAS/PI(3)K pathway via genetic alterations in EGFR, FGFR3, NTRK1, ROS1, PDGFRA, PIK3CA, RAS, MET, KIT, PTPN11, NF1, and PTEN genes ii) Cell cycle pathways via inactivating mutation/loss of TP53, CDKN2A, and RB1 genes and gain of function of CDK4, and CDK6 genes and iii) TERT promoter mutations.[10] Pediatric high-grade gliomas are unique, featuring mutations and/or copy number alterations in H3F3A, HIST1H3B, HIST1H3C, SETD2, ATRX, NF1, MYCN, PDGFRA, and BRAF genes. IDH mutations are rare and usually restricted to adolescent patients. These tumors can also harbor fusions involving BRAF, FGFR, NTRK1, NTRK2, NTRK3, and ALK genes [12-16].
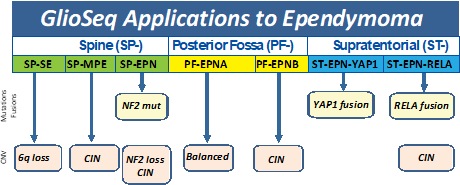
Ependymomas have been classified into molecular subgroups using a combination of genomic alterations and anatomic location along the neuraxis. Based on presence of mutations and copy number alterations, GlioSeq v3 test is able to established molecular classification in 76% of ependymomas. [17] RELA gene fusion defines a distinct clinicopathologic subtype of supratentorial ependymomas associated with chromothripsis, multiple copy number abnormalities, aggressive biologic behavior, and shorter overall survival. In contrast, supratentorial ependymomas with YAP1 gene fusion, NF2-mutated and 6q-deleted spinal ependymomas as associated with favorable clinical outcomes.[18]
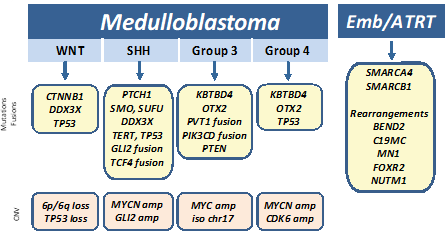
Medulloblastomas have been recently classified into four groups (WNT (wingless), SHH (sonic hedgehog), Group 3, and Group 4) based on molecular profiling and clinical outcome. Mutations in CTNNB1 and DDX3X, with or without inactivating TP53 alterations, are typically seen in Wnt-pathway medulloblastomas that are associated with a better prognosis. Tumors with PTCH1, SMO, SUFU, and TERT promoter alterations and fusions in GLI2, TCF4, and DDX3X genes characterize the SHH class medulloblastomas, and have an intermediate prognosis between Wnt and group 3/4 tumors. In contrast, mutations in KBTBD4 and OTX2, MYC amplification and fusions involving PVT1, PIK3CD, and PTEN have been described in group 3 medulloblastomas and MYCN/CDK6 amplification, mutations in KBTBD4 and OTX2, and TP53 rearrangements are more common in the group 4 medulloblastomas. The group 3 and 4 medulloblastomas are far more likely to have a poor prognosis even with therapy.[19, 20] Other types of CNS embryonal tumors, including atypical teratoid/rhabdoid tumor (ATRT) can harbor gene rearrangements in C19MC (microRNA cluster region), BEND2, MN1, CIC, and FOXR2 and mutations in SMARCA4 and SMARCB1.[21-23]
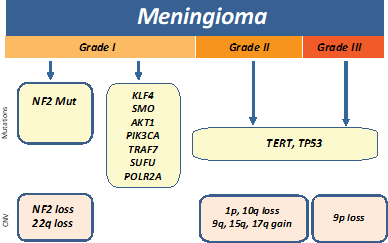
Meningiomas are classified by the WHO into benign meningiomas (grade I), atypical (grade II), and anaplastic (grade III). Inactivation of NF2 via mutation or loss of 22q is the most common early genetic alteration in meningiomas and found in ~60% of sporadic cases. Recurrent mutations in KLF4, AKT1, SMO, PIK3CA, TRAF7, SUFU, and POL2RA genes are often present in NF2-negative sporadic meningiomas.[24-26] GlioSeq v3 test allows for risk stratification by detecting genetic alterations in genes which are most often associated with benign tumors (such as TRAF7, AKT1, KLF4, SMO, PIK3CA and POLR2A) and the assessment of alterations which are associated with malignant progression such as TERT-promoter mutation and multiple copy number alterations including 1p, 10q, and 9p (CDKN2A) loss.
References
- Nikiforova, M.N., et al., Targeted next-generation sequencing panel (GlioSeq) provides comprehensive genetic profiling of central nervous system tumors. Neuro Oncol, 2016. 18(3): p. 379-87.
- Roy, S., et al., Clinical Utility of GlioSeq Next-Generation Sequencing Test in Pediatric and Young Adult Patients With Brain Tumors. J Neuropathol Exp Neurol, 2019.
- Collins, V.P., D.T. Jones, and C. Giannini, Pilocytic astrocytoma: pathology, molecular mechanisms and markers. Acta Neuropathol, 2015. 129(6): p. 775-88.
- Picca, A., G. Berzero, and M. Sanson, Current therapeutic approaches to diffuse grade II and III gliomas. Ther Adv Neurol Disord, 2018. 11: p. 1756285617752039.
- Johnson, A., et al., Comprehensive Genomic Profiling of 282 Pediatric Low- and High-Grade Gliomas Reveals Genomic Drivers, Tumor Mutational Burden, and Hypermutation Signatures. Oncologist, 2017. 22(12): p. 1478-1490.
- Gutmann, D.H., et al., Somatic neurofibromatosis type 1 (NF1) inactivation characterizes NF1-associated pilocytic astrocytoma. Genome Res, 2013. 23(3): p. 431-9.
- Ellison, D.W., et al., cIMPACT-NOW update 4: diffuse gliomas characterized by MYB, MYBL1, or FGFR1 alterations or BRAF(V600E) mutation. Acta Neuropathol, 2019. 137(4): p. 683-687.
- Ramkissoon, L.A., et al., Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. Proc Natl Acad Sci U S A, 2013. 110(20): p. 8188-93.
- Cancer Genome Atlas Research, N., et al., Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med, 2015. 372(26): p. 2481-98.
- Aldape, K., et al., Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol, 2015. 129(6): p. 829-48.
- Brat, D.J., et al., cIMPACT-NOW update 3: recommended diagnostic criteria for "Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV". Acta Neuropathol, 2018. 136(5): p. 805-810.
- Korshunov, A., et al., Integrated analysis of pediatric glioblastoma reveals a subset of biologically favorable tumors with associated molecular prognostic markers. Acta Neuropathol, 2015.
(5): p. 669-78. - Gajjar, A., et al., Pediatric Brain Tumors: Innovative Genomic Information Is Transforming the Diagnostic and Clinical Landscape. J Clin Oncol, 2015. 33(27): p. 2986-98.
- Korshunov, A., et al., H3-/IDH-wild type pediatric glioblastoma is comprised of molecularly and prognostically distinct subtypes with associated oncogenic drivers. Acta Neuropathol, 2017. 134(3): p. 507-516.
- Wu, G., et al., The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet, 2014. 46(5): p. 444-450.
- Castel, D., et al., Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol, 2015. 130(6): p. 815-27.
- Roy, S., et al., N.e., Utility of GlioSeq next-generation sequencing for molecular classification of ependymomas. Modern Pathology, 2018.
- Pajtler, K.W., et al., Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer Cell, 2015. 27(5): p. 728-43.
- Gajjar, A.J. and G.W. Robinson, Medulloblastoma-translating discoveries from the bench to the bedside. Nat Rev Clin Oncol, 2014. 11(12): p. 714-22.
- Northcott, P.A., et al., The whole-genome landscape of medulloblastoma subtypes. Nature, 2017. 547(7663): p. 311-317.
- Kleinman, C.L., et al., Fusion of TTYH1 with the C19MC microRNA cluster drives expression of a brain-specific DNMT3B isoform in the embryonal brain tumor ETMR. Nat Genet, 2014. 46(1): p. 39-44.
- Sturm, D., et al., New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs. Cell, 2016. 164(5): p. 1060-1072.
- Fruhwald, M.C., et al., Atypical teratoid/rhabdoid tumors-current concepts, advances in biology, and potential future therapies. Neuro Oncol, 2016. 18(6): p. 764-78.
- Clark, V.E., et al., Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science, 2013. 339(6123): p. 1077-80.
- Yuzawa, S., H. Nishihara, and S. Tanaka, Genetic landscape of meningioma. Brain Tumor Pathol, 2016. 33(4): p. 237-247.
- Clark, V.E., et al., Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat Genet, 2016. 48(10): p. 1253-9.
Examples of GlioSeq Reports
Specimen Requirements and Shipping Instructions
Paraffin embedded tissue sections
- Tissue should be fixed in formalin and not exposed to decalcification solution. The paraffin block should contain no less than 1-3 mm area of tumor.
- Slides should prepared by histology using a specific protocol for cutting molecular sections to avoid contamination of the tissue sections.
- One H&E and 6 unstained sections are required for most of the tests. Ten unstained sections or more are requested if the tissue is small. Please call the lab if you have questions.
- Slides should be properly labeled with a block label that matches the surgical pathology specimen number on the surgical pathology report.
- Slides should be sent ambient temperature in proper storage containers (plastic slide boxes) to protect them during shipment.
- A surgical pathology report and completed requisition must accompany all specimens.
Frozen tissue
- A minimum of 2 x 2 x 2 mm of frozen tissue is required.
- Collection date and time should be stated.
- Tissue specimen containing at least 50% of tumor cells can be either placed into cryogenic tube and snap frozen in liquid nitrogen, or placed into a tube with preservative solution provided by the Molecular & Genomic Pathology laboratory (request solution from the lab) and frozen at -20° C.
- Ship overnight on dry ice. A surgical pathology and/or cytology report and completed requisition form must accompany all specimens.
Turnaround Time
Within 10-14 business days of receipt
Billing Information
Please contact us for billing and price information.
© 2026 UPMC | University of Pittsburgh Medical Center.
Designed and maintained by Enterprise Provider Solutions.
Send questions and comments to the Web Administrators.
